Biotechnology Principles and Processes NCERT: This article discusses the Biotechnology Chapter of NCERT Class 12. Here we will discuss the Principles of Biotechnology and the tools and techniques of biotechnology. This post is the continuation of our Plus Two Botany Notes series. Biotechnology or Genetic engineering uses various types of tools and techniques such as Cloning Vectors, Restriction Endonucleases, Electrophoresis, Competent Cells etc. We will see each of it in detail. You can see the Part-2 of this article (Applications of Biotechnology) from here. The PDF version of this notes can be freely downloaded from the link given at the end of the article.
What is Biotechnology?
Biotechnology is the technology that make use of living organisms or enzymes from living organisms to produce products and processes for specific purposes useful to humans. It is the integration of natural sciences and engineering in order to achieve the application of organisms, cells, parts thereof and molecular analogues for products and services.
(1). Techniques that enabled the birth of Biotechnology:
Ø The two core techniques that enabled the birth of modern biotechnology:
(a). Genetic engineering
Ø It is the technique to alter (or manipulate) the genetic make-up (DNA/RNA) of an organism
Ø Change in genotype changes the phenotype
(b). Maintenance of sterile ambience in chemical engineering processes
Ø Only the desired organisms should grow to produce the specific product
Ø To ensure this, sterile ambience should be maintained
(2). Sexual reproduction is considered more advantageous than asexual reproduction. Why?
Ø Sexual reproduction creates genetic recombination and thus variation.
Ø Asexual reproduction does NOT create genetic recombination and variation.
Ø Thus, sexual reproduction is considered more advantageous than asexual reproduction.
(3). Who constructed the first artificial recombinant DNA?
Ø Stanley Cohen and Herbert Boyer in 1972

Ø They linked a gene encoding antibiotic resistance to the native plasmid of Salmonella typhimurium.
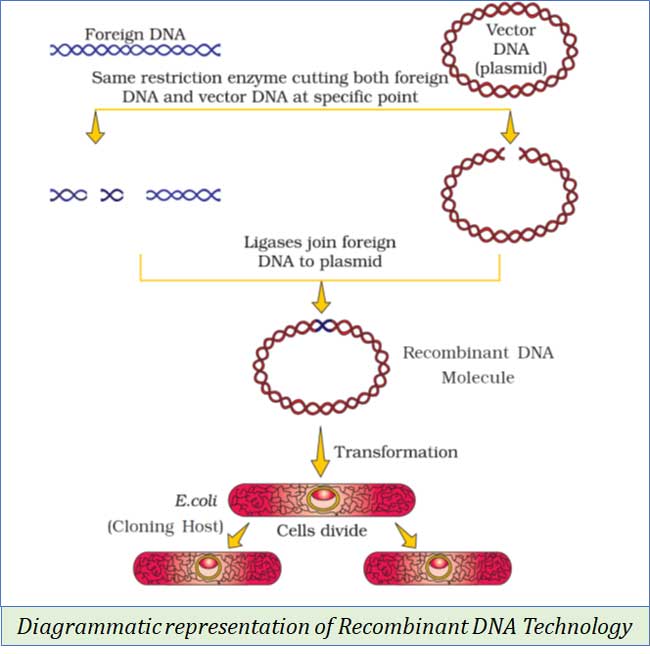
(4). Tools in Recombinant DNA Technology
(a). Restriction enzymes: Used for cutting DNA
(b). DNA Ligase: Used for linking or joining DNA molecules
(c). DNA polymerase: Used for gene cloning or for producing multiple copies of DNA
(d). Vectors: These are vehicles used to transfer a gene from one cell to another
(e). Host organism/cell: The recombinant DNA (having the desired alien DNA) can replicate inside the host organism
(5). Enzymes used in recombinant DNA technology
Ø Restriction enzymes: Used for cutting DNA
Ø DNA Ligase : Used for linking or joining DNA molecules
Ø DNA polymerase : Used for gene cloning or for making multiple copies of DNA
(6). Name the first isolated restriction endonuclease enzyme:
Ø Hind II form Haemophilus influenzae
(7). What is a recognition sequence?
Ø Each restriction enzyme recognizes a specific DNA sequence of six base pairs and cut the DNA molecule at this point.
Ø This specific base pair sequence is called recognition sequence or restriction site.
(8). Explain the defense mechanism in Escherichia coli.
Ø The restriction modification system (RM system) found in bacteria and other prokaryotic organisms provides defense against foreign DNA.
Ø It involves the action of two enzymes – Restriction Enzyme and Modification Enzyme
(a). Restriction Enzyme
Ø It recognizes the foreign DNA and cleaves it into fragments at specific points
Ø Thus, it destroys the foreign DNA
(b). Modification enzyme (Methylase)
Ø The bacterium itself will contain same base pair sequence in its genome that is recognized by the restriction enzyme.
Ø In order to prevent destruction of its own DNA by the restriction enzymes, methylase enzyme adds methyl group to one or two bases within the sequence.
Ø Thus, bacteria protect their own DNA from cleavage.
(9). Nucleases and their Classification
Ø Nucleases are enzymes which cleave nucleic acids.
Ø They cleave the phosphodiester bonds between nucleotides of nucleic acids
Ø Nucleases are of two types
(1). Exonucleases
Ø Exonucleases digest nucleic acids from the ends.
(2). Endonucleases
Ø Endonucleases make cuts at specific points within the DNA
Ø Eg. Restriction enzymes
Ø Restriction Endonuclease (REN) is a group of endonuclease enzyme.
Ø Restriction endonucleases are important tool in biotechnology
(10). Naming of Restriction Enzymes
Ø Example: EcoRI
Ø EcoRI was isolated from the bacterium Escherichia coli RY 13
Ø E –Taken from the genus name Escherichia
Ø Co – Taken from the specific epithet (species name) coli
Ø R – Taken from the name of the strain RY 13
Ø I – It was the first endonuclease isolated from the strain RY 13
(11). Action of Restriction Endonuclease
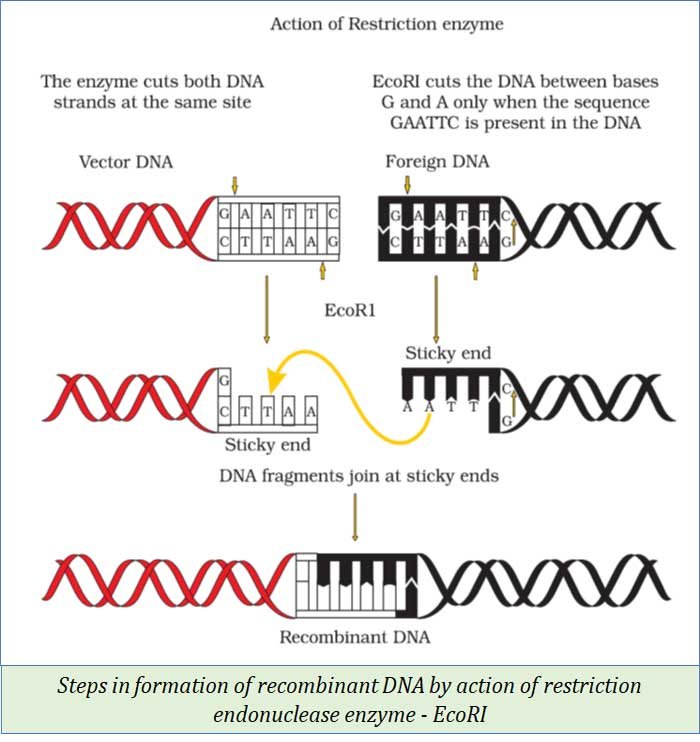
Ø Restriction endonuclease enzyme cleaves the DNA only at recognition sequence.
Ø A recognition sequence is a nucleotide sequence composed of 4, 6, or 8 nucleotides that is recognized by a restriction endonuclease.
Ø Recognition sequences are palindromic nucleotide sequence.
Ø A palindromic sequence is a base pair sequence in DNA that can be read same when the orientation of reading is same on two strands.
Ø We can read both strands either in 5’-3’ direction or in 3’-5’ direction
Ø Example for palindromic sequence: 5’ GAATTC 3’
3’ CTTAAG 5’
(12). Style of DNA cleavage
Ø Restriction enzymes cleave the DNA a little away from the centre of the palindrome site
Ø But it occurs between the same two bases on the opposite strands
Ø Cleavage results in the formation of single stranded free ends called sticky ends.
(13). GEL ELECTROPHORESIS
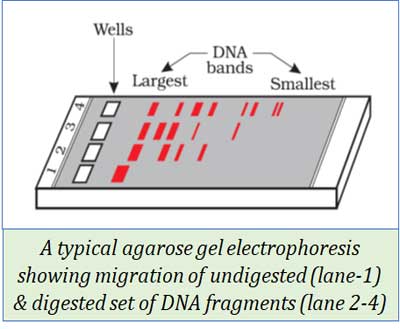
Ø Gel electrophoresis is a technique for separating DNA fragments.
Ø It is based on the size of DNA fragments.
Procedure:
Ø Cutting sample DNA into fragments by restriction endonuclease enzyme.
Ø Negatively charged DNA molecules move towards the anode under an electric field
Ø The matrix or medium used for the movement of DNA fragments is Agarose gel.
Ø Due to the sieving property of agarose gel, DNA fragments get separated according to their size
Ø Smaller fragments move farther.
Ø The separated DNA fragments are stained using Ethidium Bromide
Ø They are then visualized under UV radiation
Ø Then the DNA fragments can be seen as orange-coloured bands
Ø Elution (gel elution): The process in which the separated bands of DNA are cut out and extracted from the gel piece
(14). CLONING VECTORS
Ø Cloning vectors are vehicles used to transfer genes from one organism to another.
Ø Plasmids are generally used as cloning vectors.
Ø Types of cloning vectors: Plasmid vectors, bacteriophages (phage vectors), cosmid vectors, transposons (jumping genes)
(15). PLASMID VECTOR
Ø Plasmids are commonly used as vectors
Ø Plasmid is a circular, double stranded, autonomously replicating extra-chromosomal DNA present in bacterial cells
Artificially Reconstructed Plasmid Vectors
Ø Naturally occurring plasmids may not possess all the characteristics required for a cloning vector
Ø So, the natural plasmids are modified by inserting certain genes
Ø Example: pBR322, pU-18
Ø These were artificially reconstructed from the natural plasmid of E. coli (Col E1)
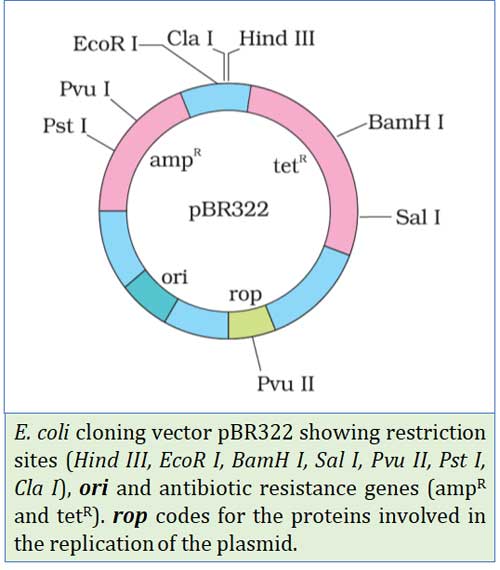
pBR322
Ø pBR322 is an artificially reconstructed plasmid vector.
Ø It was reconstructed from the natural plasmid of E. coli (Col E1) by adding (inserting) three genes
Ø Three genes inserted are:
(1). ori – Origin of replication: gene for starting the replication
(2). tetR – Gene for tetracycline (an antibiotic) resistance
(3). ampR – Gene for ampicillin (an antibiotic) resistance
Ø tetR and ampR are called selectable markers
Ø pBR322 is the first artificial cloning vector

Ø It was developed by Boliver and Rodriguez
Ø P – plasmid, B – Boliver, R – Rodriguez
(16). FEATURES OF A CLONING VECTOR
(a). Origin of replication (ori)
Ø It is the DNA sequence that initiates DNA replication
Ø It controls the copy number of the inserted DNA
Ø Any piece of DNA when linked to ori can be made to replicate within the host cell
(b). Selectable markers
Ø It is the gene which helps in identifying and eliminating non-transformants and selectively permitting the growth of transformants
Ø Transformant: A bacterial cell which contain a foreign DNA (The cells that have undergone transformation)
Ø Non-Transformant: A bacterial cell which do not contain a foreign DNA
Ø Transformation: The process by which a foreign DNA is inserted into a bacterial cell
Ø Two selectable markers in pBR322 are tetR and ampR
Ø tetR : gene for tetracycline resistance : give resistance against the antibiotic, tetracycline
Ø ampR : gene for ampicillin resistance : give resistance against the antibiotic, ampicillin
(c). Cloning site
Ø It is the recognition site where the foreign DNA can be inserted
Ø eg. BamHI, HindIII, SalI, PvuI, PvuII, PstI, EcoRI, and ClaI
(17). INSERTIONAL INACTIVATION
Ø Insertion of any foreign DNA at the cloning site of a gene in a cloning vector will cause inactivation of that gene
Ø By this method, Recombinants and Non-recombinants can be identified
Ø Non- Transformant: A cell without plasmid vector
Ø Transformant: A cell containing plasmid vector
Ø Recombinant: A cell transformed with recombinant plasmid vector
Ø Non-recombinant cell: A cell transformed with non-recombinant plasmid vector
Selection of Recombinants by Inactivation of Antibiotics
Ø When a foreign DNA is ligated at BamHI site (cloning site) of tetR gene, pBR322 will lose tetracycline resistance
Ø Non-Transformants: cannot grow on the medium containing both tetracycline and ampicillin
Ø Non-recombinants: can grow on the medium containing both tetracycline and ampicillin
Ø Recombinants: can grow on the medium containing ampicillin but not on that containing tetracycline
Selection of Recombinants by Inactivation of Enzyme
Ø β-galactosidase is a chromogenic enzyme which produces blue colour
Ø Insertion of a foreign gene into the coding sequence of β-galactosidase inactivates the gene
Ø Thus, inactivation of the gene causes the inactivation of the enzyme
Ø Such inactivated enzymes do not produce colouration
Ø Recombinants: do not show blue colouration
Ø Non-recombinants (bacterial colonies without inserted DNA): show blue colouration
(18). GENE TRANSFER
Ø Transfer of genes from one organism to another
Ø There are two methods for gene transfer
(a). Gene transfer through vectors
(b). Gene transfer without vectors / Direct gene transfer into the cell
(a). Gene Transfer Through Vectors
(i). Plasmid vectors of Agrobacterium tumifaciens
Ø Agrobacterium tumifaciens is a soil bacterium
Ø It is a pathogenic bacterium which causes crown gall (plant tumour) in dicot plants
Ø Plasmid of Agrobacterium: Ti plasmid
Ø The transferable part in Ti plasmid: T-DNA
Ø T-DNA is the tumour causing gene
Ø Ti plasmid is disarmed and modified to be used as a vector
Ø It is used to transfer foreign genes into plant cells
(ii). Retrovirus
Ø It is a virus which causes cancer in animals
Ø It is disarmed and used to transfer foreign genes into animals cells
(b). Gene Transfer without vectors / Direct Gene transfer into the cell
(i). Micro-injection
Ø Used to transfer foreign gene into animal cell
Ø Foreign gene (rDNA) is introduced into the nucleus of an animal cell using micropippettes
(ii). Gene gun / Biolistics gun
Ø Used to transfer foreign gene into plant cell
Ø Micro-particles of gold or tungsten is coated with foreign DNA
Ø Then these micro-particles are bombarded into plant cell with high velocity
(19). COMPETENT HOST
Ø The cell which is capable of taking up foreign DNA
Ø Competence: It is the ability of a cell to take up genes and plasmids from outside
Why DNA cannot pass through cell membrane?
Ø DNA is negatively charged and hydrophilic, whereas cell membrane is hydrophobic
Ø Cell membrane is mainly composed of lipids
Ø It will not allow hydrophilic molecules such as DNA to cross the cell membrane
How to make bacterial cell competent?
Ø It is done by a treatment method called Heat Shock Treatment
Heat Shock Treatment:
(1). The cell is treated with specific concentration of a divalent cation such as calcium
(2). It is done to increase the pore size in cell wall
(3). The cells are then incubated with recombinant DNA on ice
(4). Then place these cells briefly at 420C
(5). Then put them back on ice
(6). The bacterial cells now can take up recombinant DNA
So far, we have completed the discussion of Part-1 of Plus Two Botany – Biotechnology – Principles of Biotechnology. In the next article we will discuss the Applications of Biotechnology. You can Download the PDF of this note from the link given below. Hope this post was helpful for your learning. We would like to hear it from you. Please COMMENT (below ↓)
<<< Back to PLUS TWO BOTANY NOTES Page
You might also like…
@. Plus Two Biology Previous Year Question Papers
@. Plus One Biology Previous Year Question Papers
@. Model Question Papers