“The formation of DNA’s structure by Watson and Crick may turn out to be the greatest developments in the field of molecular genetics in recent years”
Linus Pauling, 1953
Purines (Adenine & Guanine) and pyrimidines (Thymine, Cytosine & Uracil) are the two classes of nucleotides which forms the nucleic acids (DNA & RNA) in the cells. Apart from the primary role of DNA and RNA as “genetic information storage”, nucleotides also serves different functions in the cells such as energy carrier (ATP and GTP), components of co-enzymes (NAD and FAD) and cellular signal transduction (cAMP and cGMP as ‘second messengers’). An ample supply of nucleotides in the cell is very essential for all the cellular processes. This post discuss the biosynthesis of Purines and Pyrimidines in an EASY but detailed way.
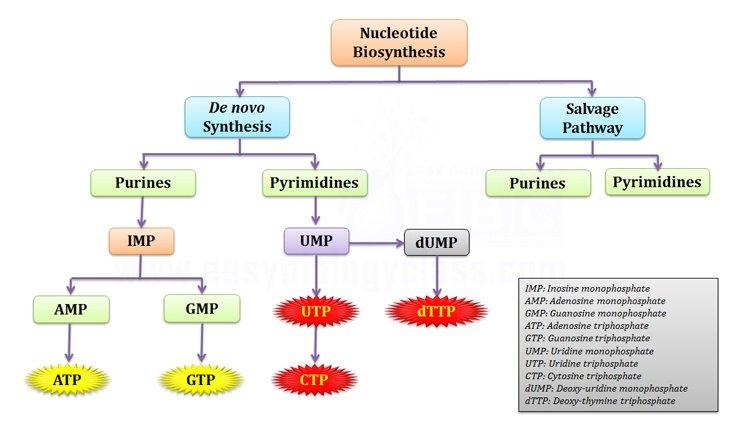
Pathways for the biosynthesis of nucleotides
Nucleotide biosynthesis in the cell can be grouped into two broad classes. (1) de-novo synthesis and (2) synthesis by salvage pathways.
I. De-novo synthesis (synthesis from scratch): it is a biochemical pathway in which nucleotides are synthesized new from simple precursor molecules.
II. Salvage pathway (recycle pathway): used to recover bases and nucleosides formed during the degradation of RNA and DNA
Learning objectives:
@. How nucleotides are synthesized in the cells?
@. How de-novo synthesis of purines & pyrimidines occurs?
@. Synthesis of IMP (precursor of Adenine and Guanine)
@. Synthesis of Adenine and Guanine from IMP
@. Synthesis of Uracil
@. Synthesis of Cytosine
@. Synthesis of deoxyribonucleotides
@. Synthesis of Thymine
@. Salvage pathways of purines and pyrimidines
I. De-novo synthesis of purines:
The purine nucleotides of nucleic acids are adenosine 5-monophosphate (AMP; adenylate) and guanosine 5-monophosphate (GMP; guanylate), containing the purine bases adenine and guanine respectively. The first idea about purine nucleotide biosynthesis in the cell was come from the study of John Buchanan (1948) by radioactive tracer studies in birds by analyzing the biochemistry of uric acid (a purine present in the excreta of birds). The detailed biosynthetic pathways of the purine biosynthesis came latter in 1950 primarily by the works of Buchanan and G. Robert Greenberg.
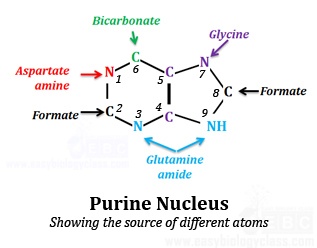 The image shows the source of different atoms in a purine skeleton identified by radio labeling studies
The image shows the source of different atoms in a purine skeleton identified by radio labeling studies
N1 is derived from amino group of Aspartate
C2 & C8 is derived from Formate
N3 & N9 is derived from amide group of Glutamine
C4, C5 & N7 is derived from Glycine
C6 is derived from HCO3- (bicarbonate)
Aspartate, Formate, Glutamine, Glycine and Bicarbonate acts as the building blocks for purine synthesis
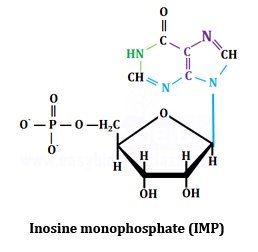
Purines (adenine and guanine) are synthesized as ribo-nucleotides (nitrogen base + ribose sugar + phosphate) rather than as free bases. Both purines are derived from a precursor namely inosine-5′-monophosphate (IMP). Thus the purine synthesis starts with IMP synthesis (See the mind map).
Synthesis of Inosine monophosphate (IMP):
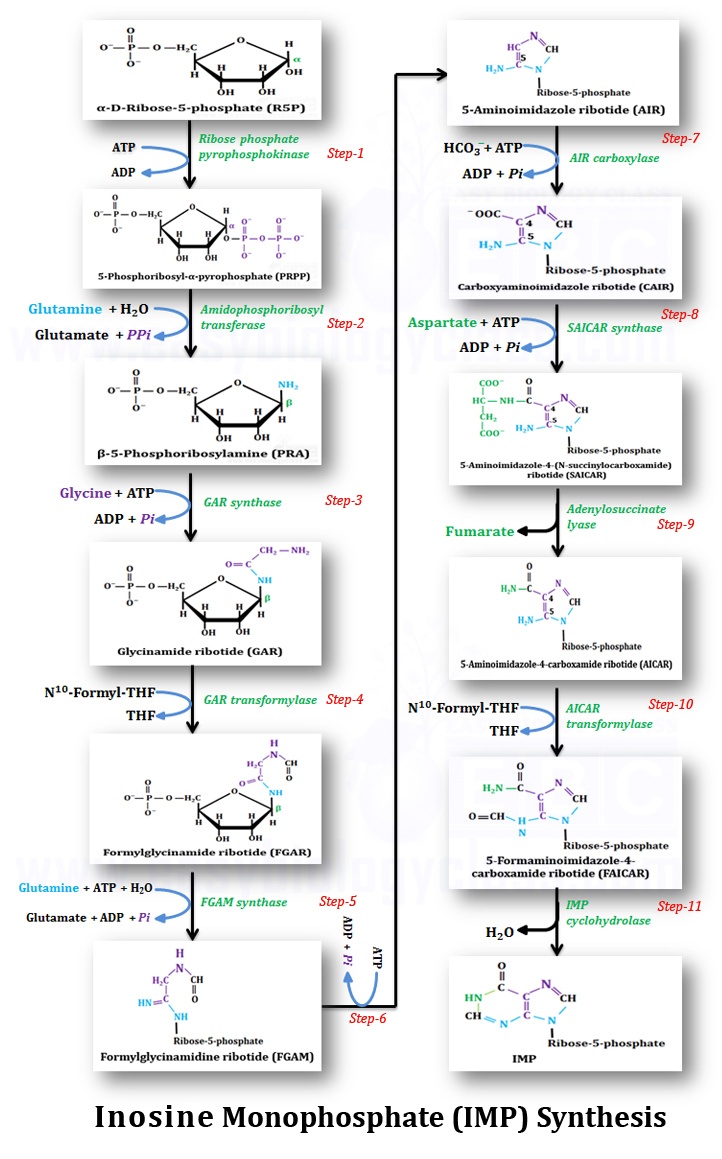
 Inosine monophosphate (IMP) is synthesized in 11 enzymatic steps from simple precursors as summarized below
Inosine monophosphate (IMP) is synthesized in 11 enzymatic steps from simple precursors as summarized below
Step-1: Ribose-5-phosphate activation and formation of PRPP): α-D-Ribose-phosphate (R5P) is activated with ATP to form 5-phosphoribosyl-α-pyrophosphate (PRPP) with the help of enzyme Ribose phosphate pyrophosphokinase. PRPP is also one of the precursors for the synthesis of pyrimidines and also the amino acids Histidine and Tryptophan.
Step-2: Acquisition of N9 atom of purine: Amide nitrogen of glutamine displaces the pyrophosphate group of PRPP and it also inverts the configuration at C1′ to form β-5-phosphoribosylamine (PRA) with the help of enzyme amidophosphoribozyl transferase. (This reaction contribute N9 atom of purine form glutamine)
Step-3: Acquisition of C4, C5 & N7 atoms of purine: Carboxylic group of glycine is combined with the amino group of β-5-phosphoribosylamine (PRA) to form glycinamide ribotide (GAR) with the help of enzyme – GAR synthetase (C4, C5, & N7 of purine are contributed by glycine)
Step-4: Acquisition of C8 atom of purine: Amino group of glycinamide ribotide (GAR) is formylated with N10-formyltetrahydrofolate and forms formylglycinamide ribotide (FGAR) with the presence of enzyme GAR transformylase. (C8 of purine is contributed by formate)
Step-5: Acquisition of N3 atom of purine: Amide nitrogen of second glutamine is added to FGAR in an ATP-dependent reaction to form formylglycinamidine ribotide (FGAM) with the help of enzyme FGAM synthetase. (N3 of purine is contributed by glutamine)
Step-6: Purine imidazole ring formation: An ATP dependent ring closing (imidazole ring formation) reaction in the presence of AIR synthetase enzyme to produce 5-aminoimidazole ribotide (AIR).
Step-7: Acquisition of C6 atom of purine: An ATP dependent carboxylation reaction of 5-aminoimidazole ribotide (AIR) with HCO3- (bicarbonate) to produce carboxyaminoimidazole ribotide (CAIR) in the presence of enzyme AIR carboxylase. (C6 of purine is contributed by HCO3-)
Step-8: Acquisition of N1 atom of purine: Aspartate is added and it forms an amide bond with C6 to form 5-aminoimidazole-4-(N-succinylocarboxamide) ribotide (SACAIR) in an ATP dependent reaction with the help of enzyme SAICAR synthetase (N1 of purine is contributed by aspartate)
Step-9: Elimination of fumarate: Fumarate group is cleaved off from SACAIR to produce 5-aminoimidazole-4-carboxamide ribotide (AICAR) with the help of enzyme- adenylosuccinate lyase.
Step-10: Acquisition of C2 atom of purine: Amino group of AICAR react with N10-formyltetrahydrofolate (formylation) to form 5-formaminoimidazole-4-carboxamide ribotide (FAICAR) with presence of enzyme AICAR transformylase. (C2 of purine ring is contributed by this N10-formyltetrahydrofolate)
Step-11: Cyclization to form IMP: In the last reaction, the larger ring of FAICAR is enzymatically closed to forms Inosine Monophosphate (IMP) with the release of a water molecule catalyzed by the enzyme IMP cyclohydrolase
.
Synthesis of Adenine and Guanine
IMP does not accumulate in the cells rather it is rapidly converted into Adenine (as AMP) and Guanine (as GMP). AMP differ from IMP in the replacement of its 6-keto group by an amino group whereas GMP differ from IMP in the presence of an amino group at C2
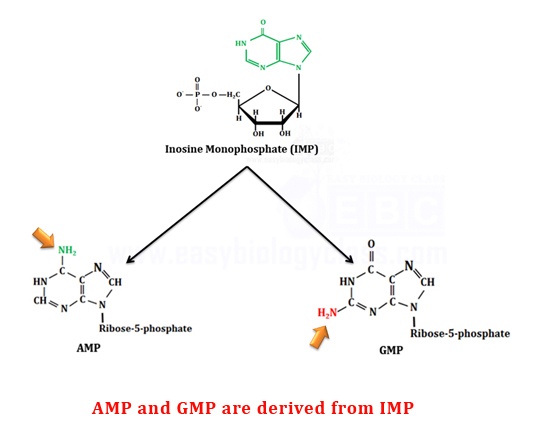
.
(a). Synthesis of AMP (Adenosine Monophosphate)
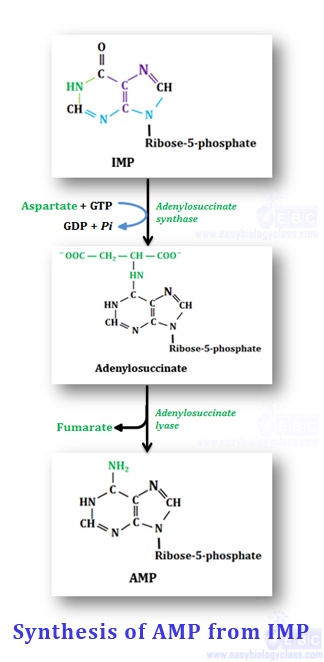 IMP is converted to AMP in two enzymatic steps
IMP is converted to AMP in two enzymatic steps
Step-1: Donation of amino group by aspartate: Amino group of aspartate is enzymatically linked to the IMP (C6 of purine) coupled with GTP hydrolysis to form adenylosuccinate with the help of enzyme- adenylosuccinate synthetase.
Step-2: Eliminates fumarate group to form AMP: Adenylosuccinate is enzymatically converted to AMP by the removal of fumarate group with the help of enzyme adenylosuccinate lyase.
(b). Synthesis of GMP (Guanosine Monophosphate)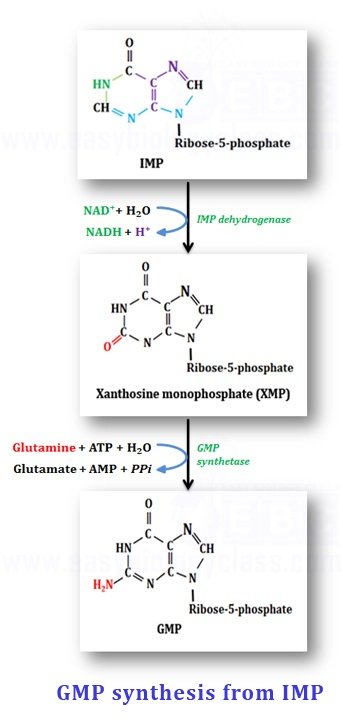
IMP is converted to GMP in two enzymatic steps
Step-1: Dehydrogenation of IMP: IMP is enzymatically dehydrogenated to form Xanthosine Monophosphate (XMP) with the enzyme IMP dehydrogenase. The H+ ions released are accepted by NAD+.
Step-2: Amidation of XMP: In the second step, XMP is amidated with the amide group from glutamine with the presence of H2O and hydrolysis of ATP yields GMP (Guanosine monophosphate); catalyzed by the enzyme GMP synthetase.
.
Synthesis of Nucleoside Diphosphates and Triphosphates
For the participation of DNA and RNA synthesis, nucleoside monophosphates and diphosphates must be converted into nucleoside triphosphates. Nucleotide diphosphates are synthesized from corresponding nucleotide monophosphate by phosphate group transfer from ATP with the help of base specific nucleoside monophosphate kinase enzyme. Similarly, nucleotide triphosphates are synthesized by the second round phosphorylation aided by ATP with the help of enzyme nucleoside diphosphate kinase.
AMP + ATP ⇌ ADP + ADP
GMP + ATP ⇌ GDP + ADP
GDP + ATP ⇌ GTP + ADP
ADP can also be converted to ATP by various energy-releasing reactions in the cells such as by oxidative phosphorylation (electron transport system of respiration), by photophosphorylation (light reaction of photosynthesis) and also by substrate level phosphorylation (as in glycolysis).
II. De-novo synthesis of Pyrimidines (Uracil, Thymine & Cytosine)
Biosynthesis of pyrimidines is simple than that of purines. Unlike purine synthesis, pyrimidines are synthesized as bases and latter it is added to ribose sugar, i.e., the ring is completed before being it is linked to ribose-5-phosphate. Following diagram shows the source of different atoms in a pyrimidine skeleton identified by radio labeling studies.
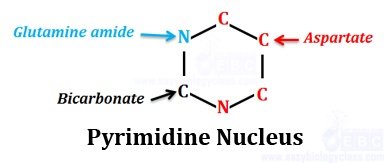 N1, C6, C5 and C4 are derived from aspartate
N1, C6, C5 and C4 are derived from aspartate
N3 is derived from glutamine
C2 is derived from HCO3- (bicarbonate)
Aspartate, Glutamine and bicarbonate contributes pyrimidine nucleus.
(a). De-novo synthesis of UMP (Uridine monophosphate)
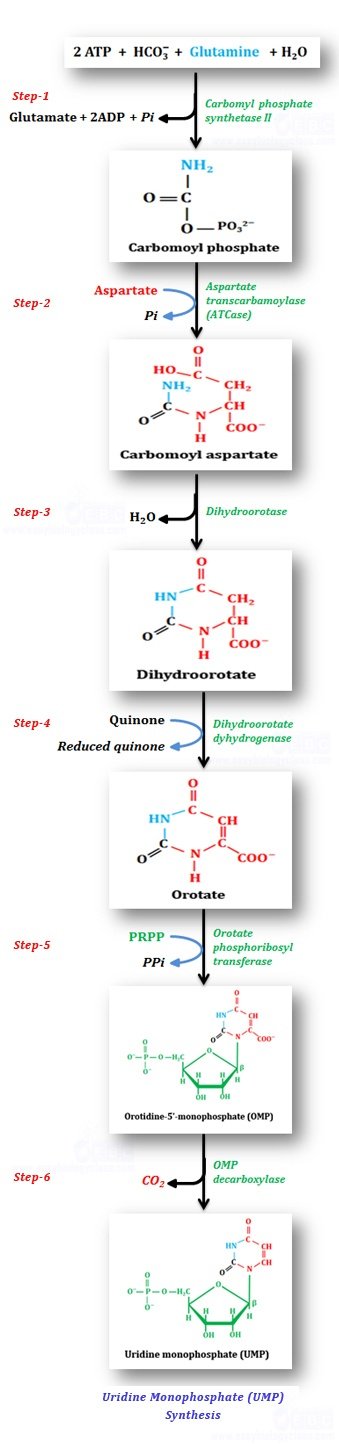 Uridine monophosphate (UMP) also acts as the precursor of CTP and dTTP). De-novo synthesis of UMP is completed in 6 enzymatic steps from simple precursors.
Uridine monophosphate (UMP) also acts as the precursor of CTP and dTTP). De-novo synthesis of UMP is completed in 6 enzymatic steps from simple precursors.
Step-1: Synthesis of carbamoyl phosphate: With the hydrolysis of two ATP molecules, bicarbonate and amide nitrogen of glutamine combine to form carbamoyl phosphate in the presence of enzyme carbamoylphosphate synthetase II.
Step-2: Synthesis of carbamoyl aspartate: Carbamoyl phosphate reacts with aspartate to yield carbamoyl aspartate catalyzed by the enzyme aspartate transcarbamoylase (ATCase).
Step-3: Ring closure & dihydroorotate formation: By the elimination (condensation reaction) of one molecule of water, the carbamoyl aspartate is converted to a ring compound – dihydroorotate catalyzed by dihydroorotase enzyme.
Step-4: Oxidation of dihydroorotate: Dihydroorotate is dehydrogenated to form orotate with the enzyme dihydroorotate dehydrogenase. (In eukaryotes, dihydroorotate dehydrogenase is located in the outer surface of inner mitochondrial membrane. All other enzymes of pyrimidine synthesis are located in the cytosol. Inhibition of dihydroorotate dehydrogenase will inhibit pyrimidine synthesis in T lymphocytes, thereby it attenuate the autoimmune disease rheumatoid arthritis. Since the enzyme is not in the cytosol, the oxidizing power required for the conversion of dihydroorate is provided by Quinone)
Step-5: Acquisition of the ribose phosphate moiety: Orotate reacts with PRPP to produce orotidine-5′-monophosphate (OMP) with the enzyme orotate phosphoribosyl transferase. The anomeric form of pyrimidine nucleotides is fixed in in the β-configuration.
Step-6: Decarboxylation to form UMP: OMP undergoes decarboxylation with assistance of enzyme OMP decarboxylase (ODCase) to form uridine monophosphate (UMP). The catalytic conversion rate of OMP decarboxylase is by a factor of 2 X 1023 over un-catalyzed reaction, making it the most catalytically proficient enzyme known to science..
Synthesis of UTP from UMP
UMP is converted to UTP in two step kinase reaction with 2 molecules of ATP
UMP + ATP ⇌ UDP + ADP
UDP + ATP ⇌ UTP + ADP.
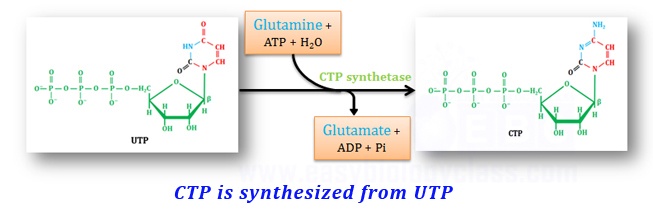
(b). Synthesis of CTP
CTP is synthesized by the amination of UTP by the enzyme CTP synthase. In animals amino group is donated by glutamine whereas in bacteria, the amino group is donated directly by ammonia.
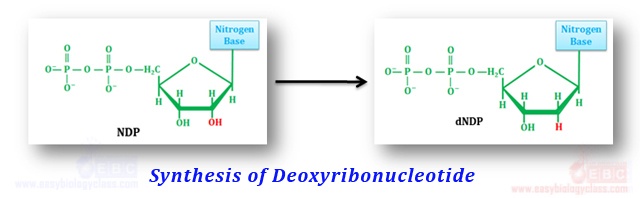
.Synthesis of Deoxyribonucleotides:
Deoxyribonucleotides are synthesized from their corresponding ribonucleotides by the reduction of ribose sugar at position C2’. Enzymes the in the formation of deoxyribonucleotides by the reduction of the corresponding ribonucleotides are called ribonucleotide reductases (RNRs). There are three classes of RNRs so far described in the living world and they all differs in their prosthetic groups. All of them replace the C2’ – OH group of ribose with – H via a free radical mechanism
 .
.
(c). Synthesis of Thymine (5-methyluracil) as dTTP:
Thymine, which is present in DNA and not in RNA, is a methylated uracil residue. Thymine in the cell is synthesized as dTTP from dUMP by methylation in four steps.
Step-1: dUTP is hydrolyzed to dUMP and PPi by the enzyme dUTP diphosphohydrolase (dUTPase)
Step-2: dUMP is then methylated to form dTMP
Step-3 & 4: dTMP is then phosphorylated with ATP in two rounds to form dTTP.
Salvage pathway of Purines
Purines can be generated in the cells during the degradation of nucleic acids through salvage pathways. Turnover of nucleic acids (particularly RNA) in most cells releases adenine, guanine, and hypoxanthine. These free purines are reconverted to their corresponding nucleotides through salvage pathways. The salvage pathways are diverse in different organism in contrast to the de-novo purine nucleotide synthetic pathway which is virtually identical in all cells.
Purines are salvaged by two different enzymes in mammals:
1. Adenine phosphoribosyltransferase (APRT) which mediates AMP formation using PRPP
Adenine + PRPP ⇌ AMP + PPi
2. Hypoxanthine–guanine phosphoribosyltransferase (HGPRT), which catalyzes the analogous reaction for both hypoxanthine and guanine
Hypoxanthine + PRPP ⇌ IMP + PPi
Guanine + PRPP ⇌ GMP + PPi
Lesch–Nyhan Syndrome (an X-linked trait and thus more common in males) is caused by the deficiency of HGPRT. This syndrome results in excessive uric acid (a purine degradation product) production which leads to neurological abnormalities, mental retardation and aggressive and destructive behavior.
.
Salvage pathway of pyramidines
Similar to purines, pyramidines are also recovered from the derivative intermediates of nucleic acids such as DNA and RNA. The recoveries of pyrimidines are catalyzed by the enzyme pyrimidine phosphoribosyltransferase which utilizes PRPP as the source of ribose-5-phsophate.
