What is Nucleotide Excision Repair (NER)?
As the name suggests it is a type of DNA repair mechanism. In nucleotide excision repair or NER, the damaged base along with a short stretch of healthy strand is removed and later the gap is refilled with correct nucleotides. Thus the NER pathway operates by ‘cut and patch’ mechanism. Even though nucleotide excision repair mechanism is present in prokaryotes and eukaryotes, the components of the pathways in both groups shows considerable variations. The prokaryotes shows relatively simple nucleotide excision repair mechanism, where as in eukaryotes, the NER pathway is quite complex with many enzymes.
Image source: wikipedia
Nucleotide Excision Repair can repair a variety of bulky lesions of the DNA such as pyrimidine dimers formed by UV irradiation and it can also remove chemically modified bases.
Classification of NER:
(1). UvrABC Endonuclease NER in Prokaryotes
(2). NER in Eukaryotes
There are two types of NER mechanism in eukaryotic cells
(a). Transcription Coupled Nucleotide Excision Repair (TC-NER)
(b). Global Genomic Nucleotide Excision Repair (GG-NER)
In this post, we will discuss only the NER mechanism of prokaryotes. The NER mechanism in prokaryotes is better known as UvrABC Endonuclease repair.
| You may also like NOTES in... | ||
|---|---|---|
| BOTANY | BIOCHEMISTRY | MOL. BIOLOGY |
| ZOOLOGY | MICROBIOLOGY | BIOSTATISTICS |
| ECOLOGY | IMMUNOLOGY | BIOTECHNOLOGY |
| GENETICS | EMBRYOLOGY | PHYSIOLOGY |
| EVOLUTION | BIOPHYSICS | BIOINFORMATICS |
Enzymes involved in NER pathway of prokaryotes:
Nucleotide excision repair pathway in prokaryotes is orchestrated by UvrABC endonuclease complex. The NER method is also assisted by DNA polymerase I and DNA ligase enzymes. UvrABC endonuclease complex is a special class of endonuclease enzyme involved in DNA repair of prokaryotes. The name is derived from the root term ultraviolet radiation, since the level of most of these enzymes will be elevated in the bacterial cells when the cells are exposed to UV light.
The UvrABC complex of E. coli consists of four enzymes. They are named as UvrA, UvrB, UvrC and UvrD (UvrD is also called as DNA helicase II). Among these Uvr enzymes, UvrB and UvrC are the actual endonucleases which cut the phosphodiester backbone of the DNA. In classical text books UvrABC complex is also called as DNA excinuclease. An excinuclease is a special type of endo-nuclease enzyme which cut two phosphodiester bonds at a time simultaneously, which is different from conventional endonucleases since usual endonucleases cut only one phosphodiester bond at a time.
NER mechanism also requires DNA polymerase I enzyme which will do the gap filling activity after the exinuclease removes the damaged bases. NER also requires DNA ligase enzyme for the final nick joining. The sequential action of these enzymes such as UvrA, UvrB, UvrC, UvrD, then DNA polymerase I and DNA ligase are essential for the orchestration of NER machinery in the bacterial cells.
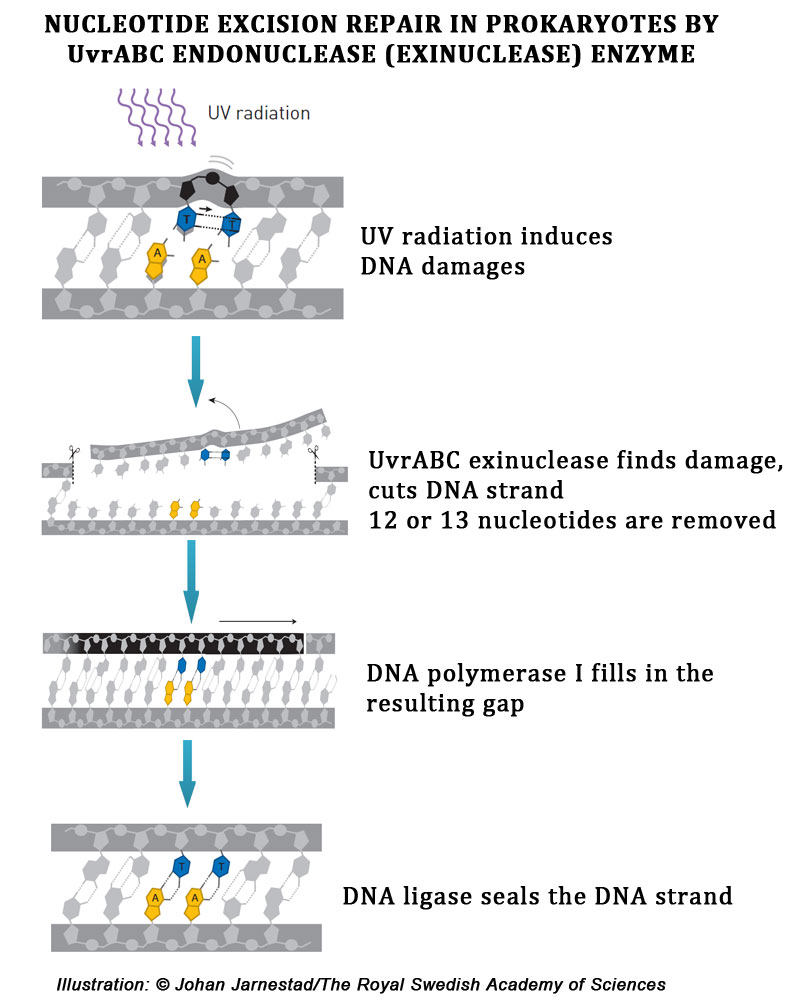 Mechanism of UvrABC complex assisted Nucleotide Excision Repair mechanism in Prokaryotes:
Mechanism of UvrABC complex assisted Nucleotide Excision Repair mechanism in Prokaryotes:
During the nucleotide excision repair mechanism in bacterial cell, first two units UvrA and one unit UvrB enzymes combined to form a heteromeric timer called UvrA2B complex (because it is with two UvrA proteins and one UvrB protein). The UvrA2B trimer complex scans the DNA for any lesions or chemical modifications in the DNA. When the UvrA2B complex finds a lesion in the DNA, it stalls there and compactly binds to the damaged region.
Then the two UvrA components are detached from the DNA, leaving the UvrB complex on the damaged portion of the DNA. The UvrB enzyme then recruits another enzyme, UvrC to the damaged area. Thus, at this state, the DNA lesion is occupied by one UvrB and one UvrC enzymes. As we mentioned previously, these two enzymes such UvrB and UvrC are the actual endonuclease in the UvrABC complex. The UvrB enzyme cleaves the phosphodiester backbone located 5 nucleotide away from the damaged base at the 3’ side of the lesion.
The UvrC enzyme cleaves the phosphodiester bond located at the 8 nucleotide away from the damaged base at the 5’ side of the lesion. This double cleavage at two points generates a 12 to 13 nucleotide long strand called 12-mer or 13-mer depending upon the number of nucleotides. After the cleavage, the UvrB enzyme is detached from the DNA. Then, to the UcrC DNA complex, a helicase enzyme called DNA helicase II or UvrD binds. Helicase enzyme displaces the 13-mer strand of the DNA along with the damaged base from the DNA duplex.
After the complete removal of the damaged portion of DNA, the UvrC also leaves the DNA. Thus, the damaged base along with a short stretch of healthy nucleotides were removed from the DNA and leaving this portion as the single stranded region. The single stranded portion of DNA will be recognized by DNA polymerase I enzyme and binds to it.
DNA polymerase I will refill the gap with correct nucleotides by consulting the nucleotide sequence in the complementary strand. Finally the DNA ligase enzyme reseals the nick formed by the addition of new nucleotides. With this the damaged portion of the DNA is removed and the original nucleotide composition is regained.
| You may also like... | ||
|---|---|---|
| NOTES | QUESTION BANK | COMPETITIVE EXAMS. |
| PPTs | UNIVERSITY EXAMS | DIFFERENCE BETWEEN.. |
| MCQs | PLUS ONE BIOLOGY | NEWS & JOBS |
| MOCK TESTS | PLUS TWO BIOLOGY | PRACTICAL |
Key points:
Ø NER or nucleotide excision repair removes the damaged bases from the DNA along with a short stretch of healthy strand and repairs it
Ø NER Pathway is present in prokaryotes and eukaryotes
Ø NER pathway in prokaryotes is facilitated by UvrABC endonuclease complex
Ø DNA polymerase I and DNA ligase enzyme are also involved in NER
Ø UvrABC complex consists of prokaryotes four enzymes
Ø They are UvrA, UvrB, UvrC and UvrD
Ø Two UvrA and one UvrB binds together to form UvrA2B trimer complex
Ø The UvrA2B complex recognize the DNA lesion and binds to it
Ø UvrB and UvrC enzyme cut the damaged strand and produce a 12-13 nucleotide long strand (12-mer or 13-mer)
Ø 12-mer or 13-mer is removed from the DNA duplex by the activity of DNA helicase II
Ø DNA polymerase refill the gap of DNA with correct nucleotides
Ø Finally DNA ligase enzyme rejoin the nick
<<Back to Molecular Biology Notes
