CSIR/JRF/NET: Life Sciences, December 2014 (II):
Part B (Original Questions)
New: NET Life Sciences Dec 2015 Question Paper with Detailed Answer key
(1). Chirality of DNA is due to:
a. The bases
b. Base stacking
c. Hydrogen bonds between bases
d. Deoxyribose
Ans: d – Deoxyribose
Biomolecules like DNA, proteins, amino acids and sugars are chiral molecules.
…
(2). Proton motive force during oxidative phosphorylation is generated in mitochondria by:
a. Exchanging protons for sodium ions
b. Pumping protons out into inter-membrane space
c. Pumping hydroxyl ions into the mitochondria
d. Hydrolysis of ATP
Ans: c – Pumping protons out into inter-membrane space
During Electron Transport System (ETS) in the inner mitochondrial membrane, protons are pumped out of the mitochondrial matrix in to peri-mitochondrial space. This creates a concentration gradient of H+ ions between mitochondrial matrix and peri-mitochondrial space to have a proton-motive force. This proton-motive force will be later utilized by the ATP synthase enzyme to synthesize ATP. In chloroplasts, during light reaction of photosynthesis, the proton-motive force is created between inner thylakoid space and chloroplast matrix. Here also this proton-motive force will be later used by ATP synthase to produce ATP.
(For details: Voet and Voet, 2012, Fundamentals of Biochemistry, Ed. 4, Chapter 18, Electron Transport and Oxidative Phosphorylation, Page: 587)
Chemiosmotic theory: (proposed by Peter Mitchel, 1961): Free energy of electron transport is conserved by pumping H+ from the mitochondrial matrix to the inter-membrane space to create an electrochemical H+ gradient across the inner mitochondrial membrane. The electrochemical potential of this gradient is harnessed to synthesize ATP
(For details: Voet and Voet, 2012, Fundamentals of Biochemistry, Ed. 4, Chapter 18, Electron Transport and Oxidative Phosphorylation, Page: 604)
…
(3). In proteins, hydrogen bonds forms as follows: Donor (D)-H— Acceptor (A). Hydrogen bond is more favourable if the angle between D-H and A is:
a. <90º
b. 180º
c. >180º
d. 120º
Ans: b – 180º
Hydrogen bond: A very weak bond formed by the electrostatic attraction between hydrogen which is covalently bonded to an electro negative atom, such as oxygen or nitrogen, to other electro negative atom. Hydrogen bond is most favourable and strong when all the three bonded atoms are in a straight line (180º). When hydrogen bonds are structurally constrained, the geometry will loose and the bonds become weaker.
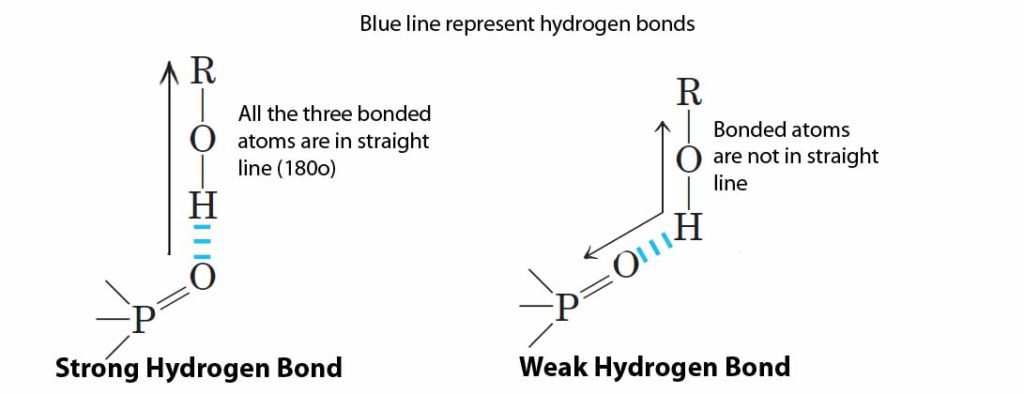
(For details: Lehninger, 2004, Principles of Biochemistry, Ed.4, Chapter 2: Water, Page: 50)
…
(4). Reaction products inhibit catalysis in enzymes by:
a. Covalent binding to the enzyme
b. Altering the enzyme structure
c. Occupying the active site
d. Form a complex with the substrate
Ans: b- Altering the enzyme structure
Feed-back inhibition: An allosteric enzymatic activity regulation mechanism in the cell to regulate metabolic pathways with more than one steps and includes more than one enzyme. Usually the first enzyme in these metabolic pathways will be an allosteric enzyme. Allosteric enzymes have “other sites” for the binding of modulators. Binding of modulators may stimulate or inhibit the activity of allosteric enzyme temporarily for short period of time. In feed-back inhibition, the end product of the metabolic pathway act as the negative modulator which binds to the allosteric site of the first enzyme and thereby making it unable to bind to the substrate and ultimately halts the metabolic pathway for a brief period of time until all the end products are consumed. The modulator only binds non-covalently (temporarily) to the allosteric enzyme.
(For details: Lehninger, 2004, Principles of Biochemistry, Ed.4, Chapter 6: Enzymes, Page: 225)
…
(5). Which of the following statements regarding membrane transport is FALSE?
a. Polar and charged solutes will not cross cell membranes effectively without specific protein carriers.
b. Each protein carrier will only bind and transport one (or a few very similar type of solute
c. Sugars such as glucose are always transported by active transport rather than by facilitated diffusion carriers
d. Ions are typically transported by special proteins that form membrane channels
Ans: c – Sugars such as glucose are always transported by active transport rather than by facilitated diffusion carriers
Due to the non-polar lipid components, polar and charged molecules will not transport directly through the plasma membrane. They require specific transporters which may be operated actively (using ATP, working against concentration gradient) or passively (no energy is required, work only along concentration gradient). The protein carriers or transporters in the membrane are highly specific and they usually allow the transport of one or very few similar type of solutes. Charged ions like Na+, K+ and Cl- etc. are transported across the membrane only through specific channels, which may be voltage gated or ligand gated.
For the transportation of glucose across plasma membrane, cell may use facilitated diffusion or active transport depending upon the specific needs and cell type. For example sugars are transported through active transport from gut to the intestinal epithelial cells where as in the case of RBC glucose is transported by facilitated diffusion. Whether active or facilitated diffusion, the glucose molecule cannot be passed directly through the membrane. They require specific transporters called Glucose Transporters (GLUTs). GLUTs are a class of membrane proteins that facilitate the transport of glucose across plasma membrane. So far 13 different types of GLUT proteins have been identified among which GLUT1, GLUT2, GLUT3 and GLUT4 are well characterized.
GLUT-1: abundant in fetal tissues, in adults present in erythrocytes.
GLUT-2: A bidirectional glucose transporter, expressed in liver, pancreatic beta cells and renal tubular cells.
GLUT-3: Glucose transporter of neurons and placenta
GLUT-4: Glucose transporter of adipose tissue, skeletal muscles and cardiac muscles
(For details: Lehninger, 2004: Principles of Biochemistry, Ed.4, Chapter 11: Biological membrane and Transport, Page: 393)
…
(6). What will happen if histones are depleted from a metaphase chromosome and viewed under a transmission electron microscope?
a. 30 nm chromatin fibres will be observed
b. 10 nm chromatin fibres will be observed
c. A scaffold and a huge number of loops of DNA fibres will be observed
d. A huge number of loops of DNA fibres without scaffold will be observed
Ans: c – A scaffold and a huge number of loops of DNA fibres will be observed
Maximum condensation of chromosome in a cell can be visualized in the metaphase phase of cell division. The metaphase chromosome, actually consists of two sister chromatids connected by a complex called kinetochore at the centromere, will be having an average thickness of 1400 nm (700 nm to each sister chromatids). The formation of chromosome during cell division is by the condensation of genetic material (DNA) with a variety of proteins generally classified as histones and non-histones. The individual DNA double helix will be having a diameter of 2 nm. The 2 nm DNA helix coils around a histone octamer (2 each of H2A, H2B, H3 and H4) to form a “bead on string” like structure having 11 nm thickness. The “beads” are actually the nucleosome particles. Each histone octamer will be surrounded by 1.65 turns of DNA with 146 to 147 nucleotides. The 11 nm “nucleosome beads on string” further coil around with the help of another histone called H1 (linker histone) to form a 30 nm solenoid model. In order to have higher level of coiling, the 30 nm fibre loops around a cellular protein scaffold. The protein scaffold consists mainly of topoisomerase II and other non-histone proteins. Looping 30 nm fibre around the scaffold create 300 nm to 700 nm metaphase sister chromosomes. Chromosome binds to scaffold proteins at specific regions called Scaffold Attachment Region (SARs).
In the question, only histone proteins are depleted which means that scaffold will not be affected. Thus the option ‘c’, i.e., a scaffold and large number of loops of DNA fibres will be observed under transmission electron microscope
…
(7). In chloroplast, the site of coupled oxidation- reduction reactions is the:
a. Outer membrane
b. Inner membrane
c. Thylakoid space
d. Stromal space
Ans: c – Thylakoid space
(For details: Stryer, 2006, Biochemistry, Ed.6, Chapter 19, Light reaction and Photosynthesis, Page: 543)
…
(8). Which of the following statements about meiosis is NOT true?
a. Kinetochores of sister chromatids attach to opposite poles in Meiosis I
b. Kinetochores of sister chromatids attach to opposite poles in Meiosis II
c. Chiasma is formed in Prophase I
d. Homologous chromosomes are segregated in Meiosis I
Ans: a – Kinetochores of sister chromatids attach to opposite poles in Meiosis I
The homologous chromosomes pairs (prophase I) and they separate to two opposite poles after the completion of Meiosis I. Splitting of kinetochore will not occurs during meiosis I (kinetochore: a protein complex which holds the two sister chromatids together and to which the spindle fibers attach during cell division). Splitting of kinetochore and separation of sister chromatids takes place only during meiosis II
…
(9). Leader sequence in some of the protozoan parasites is transcribed elsewhere in the parasite genome and gets joined with several transcripts to make the functional RNA. The joining of the two transcripts occur by the process of:
a. Alternate splicing
b. Trans splicing
c. Ligation
d. RNA editing
Ans: b – Trans splicing
Trans-splicing: A type of RNA splicing found in some eukaryotes where exons of two separate primary transcripts were joined end to end (ligated) to form a single functional mRNA. The usual splicing where the introns are removed and the remaining exons are joined with in a primary transcript is called cis- splicing. Trans splicing is responsible for the oncogenic properties of some transcripts (fusion transcripts). The process of trans-splicing was first discovered in Trypnosoma and later shown to occur in Caenorhabditis elegans and other nematodes.
RNA editing: Process of changing the specific nucleotide sequence of RNA after it has been produced by transcription. The post transcriptional modifications should not be confused with RNA editing. The post transcriptional modifications like splicing, 5’ capping, 3’ polyadenylation are not considered as RNA editing.
Alternate splicing: A method of splicing, by which single gene can produce multiple proteins with different functions. In alternate splicing the differential inclusion or exclusion of mRNA region create different mRNA sequences. Alternate splicing increases the diversity of protein coded by a single gene. Alternate splicing is a universal phenomenon and in human about 40 – 60% genes show alternate splicing.
…
(10). Small nuclear RNAs used to process and chemically modify rRNAs are called
a. Sca RNAs
b. Si RNAs
c. Sno RNAs
d. Sn RNAs
Ans: c – SnoRNA
SnoRNA: Small nucleolar RNA, they are the components of SnoRNP (small nucleolar ribonucleoproteins). snoRNP consists of small nucleolar RNA (snoRNA) packed with specific proteins. SnoRNA helps in the processing and modification of pre-rRNA
(For details: Gerald Karp, 2010, Cell and Molecular Biology: Concepts and Experiments, Ed.6, Chapter 11, Gene expression from Transcription to Translation, Page 432)
Sca RNA: Small Cajal body specific RNA, a class of small nuclear RNA (snRNA) localized to Cajal bodies (CBs).
Cajal body: They are non membrane bound special small sub-organelles present in the nucleus of actively dividing cells and they mainly consist of proteins and scaRNA. First described as nucleolar accessory bodies by Santiago Ramon Cajal. Functions of Cajal body (CBs) may be (1) SnRNPs biogenesis, (2) maturation and recycling of histone mRNA and (3) telomere maintenance. Addition of nucleotide to telomere with the help of RNA requires the helps of Cajal bodies.
SiRNA: Small interfering RNA. also called as silencing RNA, participate in RNAi (RNA interference) to interfere the expression of a targeted gene. Si RNA initiates mRNA breakdown before translation and thus protein synthesis is prevented.
